Keeping the Faith
UCSB researchers develop novel technique to determine how faithfully DNA modifications are
copied from one cell generation to the next

By Andrew Masuda, UCSB Engineering
Every human starts out as a single cell, a fertilized egg, which then divides and develops into
various cell types, such as skin cells, liver cells, and blood cells. Although these cell types look
and function differently, they contain exactly the same DNA. But, how can the same genetic
code be used to build more than two hundred different cell types in a human body?
Researchers have shown that a cell’s function comes down to the thousands of genes that are
present in a person’s DNA sequence, and which genes are active or inactive that enable it to
perform unique tasks.
Several kinds of DNA modifications influence which genes are switched on or off. A key
modification is DNA methylation, a process by which chemical structures, called methyl groups,
are added to cytosine (one of the four nucleic acids in DNA and RNA) to form 5mC. Methylation
is critical for maintaining stable repression of target genes, establishing parent-specific
expression of genes, and maintaining specific cell-type identity. Once methylation occurs, the
modification can be memorized and passed on during each round of cell division by an enzyme,
DNA-methyltransferase 1 (DNMT1), which helps to ensure that a cell’s identity is correctly
inherited in future cell generations. Epigenetics is the field of study that investigates these
altered states that are inherited without changes to the DNA sequence.
Siddarth Dey, an associate professor of chemical engineering and bioengineering at UC Santa
Barbara, and members of his research group investigate how different epigenetic factors
regulate gene expression. They have honed in on DNA methylation, seeking a better
understanding of how DNMT1 works.
“When DNA is copied, it has extremely high fidelity, which is why all of our DNA is the same,
and there are relatively few mutations between cells,” said Dey. “However, it’s a different story
with DNA methylation. DNMT1 does not copy DNA methylation as faithfully as DNA. We want to
know why, and what factors potentially affect this process.”
Researchers in the Dey lab developed a new technique to measure how faithfully DNA
methylation is copied from one cell generation to the next. Their novel method overcame a
limitation associated with a widely used technique, which provided bulk measurements for a
specific location in the genome.
“One cell type might copy methylation with high accuracy, but another nearby cell in the same
tissue might not show the same characteristics,” explained Dey. “The existing method provided
an average, but you would have no idea that these two different cell types copied DNA
methylation with different fidelity. We wanted to discover if this was the case.”
“Our technique is scalable down to a single cell, which provides insight that you would not be
able to gain in bulk,” added Alex Chialastri, who completed his PhD in chemical engineering at
UCSB in 2022 and spent nearly two years working on the project as a member of Dey’s
research group. “Showing the dynamics of individual cells provides great insight and allows us
to better understand how biological systems work, which is a key step if we want to engineer
these systems.”
Chialastri was the lead-author of a paper published recently in Nature Structural & Molecular Biology, in which they discussed their new technology and key findings, including identifying data-supported rules for when DNMT1 copies DNA methylation more faithfully.
“We found that a region that begins with high amounts of DNA methylation tends to be copied
much more faithfully than regions with lower levels,” said Chialastri. “Essentially, the density of
DNA methylation at a location was highly correlated with how efficiently the modification was
copied.”
They also found a correlation between DNA methylation fidelity and chemical modifications to
histones, a type of protein that binds to DNA and helps control gene activity.
“This finding is significant because histone modifications have been shown to play a role in the
activation and repression of genes,” explained Dey. “These observations provide additional
evidence of how histone marks and DNA methylation work in concert to suppress or turn on
genes.”
The DNA Sequencing Method
DNA sequencing is a technique for determining the order of the four chemical building blocks,
called bases, that make up a DNA molecule. The sequence tells scientists the kind of genetic
information that is carried in a particular DNA segment. In the DNA double helix, the four
chemical bases always bond with the same partner to form base pairs. Adenine (A) always pairs with thymine (T), and cytosine (C) always pairs with guanine (G). The pairing provides the basis for the mechanism by which DNA molecules are copied when cells divide, and the pairing also underlies the method by which DNA sequencing experiments are done.
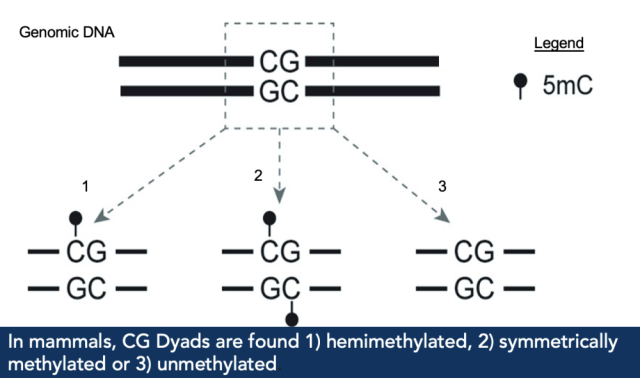
In order to quantify the genome-wide methylation status of cytosines at a single-cell level, the
Dey group developed a technique called Dyad-sequencing. As cytosine methylation in
mammals occurs in a “CG” sequence context (or a dinucleotide or “dyad”), the reverse DNA
sequence also has a “CG” sequence, with DNMT1 copying methylation from a “C” on one “CG”
to the “C” on the reverse DNA sequence. In Dyad-sequencing, the Dey lab used an enzyme,
MSPJ1, and other conversion reactions to detect the methylation status of the two C’s in a “CG”
dyad.
“The enzyme allows us to detect 5mC on one DNA strand,” explained Chialastri. “And the other
conversion reaction then allows us to infer if the cytosine on the reverse DNA strand is
methylated or not.”
In addition to DNA methylation, researchers have also studied another type of DNA modification
in mammals, called DNA hydroxymethylation (5hmC), which involves the methyl group in DNA
methylation being replaced by a hydroxyl group. 5hmC is potentially important in epigenetics,
because the hydroxymethyl group plays a role in gene expression and scientists are
investigating its possible role in aging and other diseases.
“Our technique allows us to quantify both 5mC and 5hmC from a single cell,” said Dey. “Before
this, you could measure one or the other, but not both from an individual cell.”
The team has already applied the technique to further examine different stages of early
embryogenesis, which is the formation and development of a fertilized egg early on in
pregnancy. They are also now collaborating with other scientists to understand how DNA
methylation profiles change as animals age. The method is also being used to study plants, because the technique can be applied to investigate any system in which an organism exhibits DNA methylation.
“The thing that I am most proud of about this work is that it wasn’t a one-off effort,” said
Chialastri, adding that the lab already added RNA detection to this technique. “We created a
tool that can be built upon to make other measurements by sequencing.”
“One of the things that I enjoy the most about being a scientist is discovering something new
about nature and learning how it works,” added Dey. “This is what I find exciting about building
new tools to make previously unidentified observations that tell you a little bit more about how
systems work at the molecular level.”
